
- #Texmacs help how to
- #Texmacs help install
- #Texmacs help full
- #Texmacs help software
- #Texmacs help free
which help you to produce professionally looking documents.
#Texmacs help free
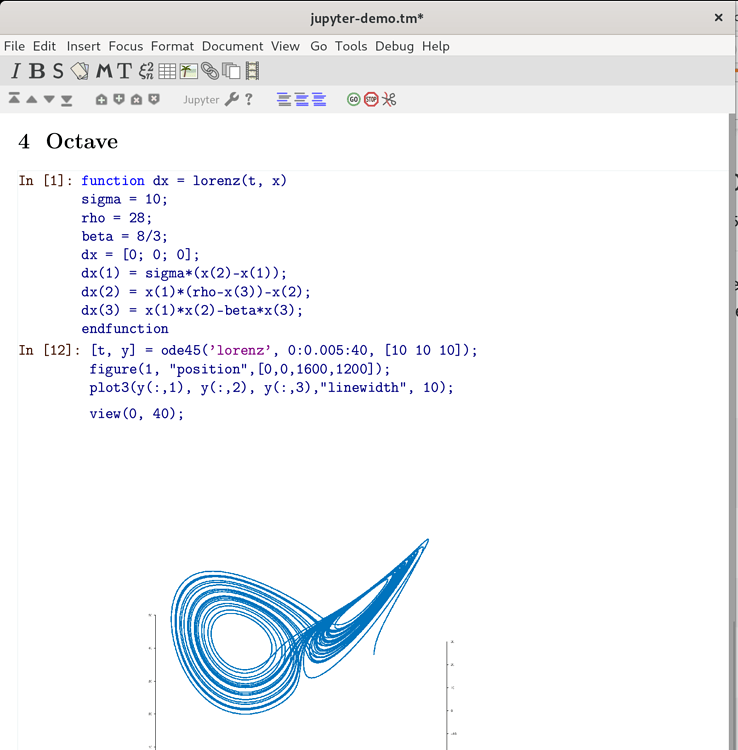
In exchange for this little initial effort, you can start writing formulas or equations more smoothly, seeing them appear on the screen as they are written (Figure 2 ). GNU TeXmacs is a free scientific text editor, which was both inspired by TeX and GNU Emacs.
#Texmacs help how to
How to contribute Documentation on how to interface TeXmacs with an extern application can be browsed in the Help->Interfacing menu or online. TeXmacs requires an understanding of basic principles that cannot be traced back to the intuitive experience of the user familiar with Word or LaTeX.
#Texmacs help install
And do NOT install PariGP using Homebrew. GSM6008689_8843_09_20200710_Clariom_S_Human_.CEL.gz For GNU TeXmacs 1.99.12 on macOS, please add /Applications/PariGP.app/Contents/Resources/bin/ to your PATH.
#Texmacs help full
Table truncated, full table size 901 Kbytes. TexMACS Medium has been developed based on the clinical TexMACS GMP Medium (170- 076-306). It is produced without animal-derived components but contains pre-selected human serum albumin, stable glutamine, and phenol red. During analysis, the genome version hg38 (Homo sapiens) and the annotation file Clariom_S_Human.r1.1.transcript.csv were applied.Įxpression of human CD8+ T cells 21 d after BCL11B KO or control treatment TexMACS Medium has a defined formulation enabling reproducible application in human and mouse cell culture. The parameter "Pos vs Neg AUC Threshold" was set to 0.7. Expression data on gene level were generated using " Gene Level - SST-RMA" as summarization method. CEL files were processed using the Transcriptome Analysis Console 4.0.2.15 (Affymetrix / Applied Biosystems by Thermo Fisher Scientific). i have taken the piecture that one can find on the page. Gene expression data of BCL11B KO CD8+ T cells after 21 d Any text editing program will do, but a specialized editor will facilitate running (La)TeX and its support programs (BibTeX, MakeIndex, etc.). RNA was processed for labeling according to Affymetrix standard workflows by Atlas Biolabs GmbH, Berlin, Germany.Īrrays were hybridized according to Affymetrix standard workflows by Atlas Biolabs GmbH, Berlin, Germany.Īrrays were scanned according to Affymetrix standard workflows by Atlas Biolabs GmbH, Berlin, Germany. TotalRNA was extracted from cells using TRIzol (Invitrogen). Furthermore, cells were re-stimulated with 2 µl/ml TransAct on d 14. Date: Fri, 00:17:35 +0100 hi javier and thanks for your mail. On d 0, two days after mock or BCL11B KO electroporation, cells were activated with 10 µl/ml TransAct (Miltenyi).
Both cytokines were re-plenished every second day. Gnu texmacs is a free scientific text editor, which was both inspired by tex and gnu emacs. Further cultivation did not differ between the experimental conditions.Ĭells were cultivated in TexMACS medium (Miltenyi) which was supplemented with antibiotics as well as with 155 U/ml IL-7 and 290 U/ml IL-15.
#Texmacs help software
00009 * If you don't have this file, write to the Free Software Foundation, Inc., 00010 * 59 Temple Place - Suite 330, Boston, MA 02111-1307, USA.GEO help: Mouse over screen elements for information.Īfter isolation of CD8+ T cells from Buffy Coat PBMCs, the cells were either directly cultivated in supplemented medium (non-treated condition, nt) or electroporated with either unspecific control gRNA-Cas9 (mock electroporated condition, mock) or with BCL11B-targeting gRNA-Cas9 (BCL11B knock-out condition, BCL11B KO). See the file $TEXMACS_PATH/LICENSE for more details. Alternatively you can use bibliography styles which do not start with tm-in which case I think TeXmacs will use the external bibtex program to process the bibliography and you can then customise his behaviour much like you do in TeX.


 0 kommentar(er)
0 kommentar(er)
